Deep Learning in Neuroscience: Connectomics and Why it Matters (Part 1 of 5)
Deep Learning in Neuroscience is a 5-part journey where you will learn about a fascinating and ambitious endeavor in neuroscience called connectomics, how deep learning models are solving practical challenges in computational neuroscience, and a few optimization techniques in PyTorch with a deep dive on implementing quantization on a research cluster.
This series aims to tell my friends and the world about a corner of neuroscience I think is super cool! I tried to write in an accessible manner so more people can share in the joy of learning about science! Where there are technical concepts, I try to follow up each technical sentence with simpler explanations and parallels to the real world.
Why Connectomics?
Imagine a world where we have access to a complete 3D diagram of all the trillions of neural connections in the human brain at the microscopic level. Research scientists are able to compare the 3D map of a healthy brain to a diseased one, leading them to uncover the physical mechanisms of psychiatric diseases. Assisted by a complete picture of the wiring in the visual cortex, neurosurgeons and engineers begin to develop a new microscopic surgical technique to restore sight to people with low vision. Knowing the structure of the brain, computer scientists begin to model their designs on the brain's architecture, making breakthroughs in building faster, more efficient, and more accurate deep learning models.
This is the world that the field of connectomics is working towards. More specifically, connectomics is a field within neuroscience that endeavors to build a connectome, which refers to the complete rendering of the neural circuit synaptic connectivity in a brain.[1] In layman's terms, research scientists are working hard so that one day, we can have a complete 3D map of all the neural connections in the human brain.
Even though the field of neuroscience has made significant progress in mapping the different regions of the brain, we still do not have a complete 3D map of the neural connections in the brain. This parallels how before the completion of the Human Genome Project in 2003, we did not have a complete genome sequence of a human being.
The undertaking of the Human Genome Project required a large upfront investment but has led to advances in the diagnosis and treatment of cancer - not to mention that every dollar invested in the project returned $141 in the US economy according to this study. Similarly, connectomics is an extremely resource-intensive project, requiring an even larger investment and the collaboration of scientists from biology, computer science, and neuroscience. However, it has the real potential to advance medicine and computer science.
Big Data Challenges of Connectomics
According to Prof. Jeff Lichtman et al. in "The Big Data Challenges of Connectomics" published in Nature (2014), the field of connectomics currently faces three main technical obstacles:
- Faster image acquisition
- Faster image processing
- Building better statistical and analytic tools
Challenge #1: Image Acquisition
In the context of connectomics, image acquisition is the process by which a cube of brain tissue is cut into thousands of thin slices and each slice is put into an electron microscope, generating a high-resolution 2D image.
At 2014 computing and image acquisition speeds, they estimate that it would take 1,000 years to map the cerebral cortex of a rat, and over 10 million years to complete a map of every synapse in the human brain.[2]
Figure 1. Carl Zeiss MultiSEM 505 in action at the Lichtman lab. Source: Jony Hu.[3]
Since then, developments in electron microscopy technology and an automated tissue slicing set-up developed in the Lichtman Lab have substantially sped up the image acquisition process. For instance, acquiring the 3D image of 1 mm3 of brain tissue used to take 6 years, but with the acquisition of a new imaging microscope from Carl Zeiss (see figure 1), it only takes one month now.[2:1] For scale, the volume of an apple seed is roughly 50 to 65 mm3.[4]
Challenge #2: Image Processing
Having acquired the images, the main bottleneck in connectomics is now processing the images. Indeed, Professor Lichtman et al. outline that "a number of analytic problems stand between the raw acquired digitized images and having access to this data in a useful form."[2:2] In other words, the problem is how do we get from a bunch of 2D image slices of the brain to a complete, accurate 3D reconstruction of all the connections in the brain?

Figure 2. Source: Lichtman Lab (Harvard University), Connectomics Team (Google)
The first problem within image processing is accuracy.[1:1] To understand the importance of accuracy, it is essential to understand how to get from the raw 2D images to the 3D map. The image above shows the end-to-end process.
- You start with a bundle of superimposed 2D black-and-white images (also known as a tensor). You feed the bundle of images into a computer vision model - think of it as a computer program that you have trained to color in images in a very particular way. Specifically, the computer vision model identifies each unique cell in the first image of the bundle and then colors in the boundaries of each cell with a different color. This task is called image segmentation.
- Moving on to the second image (the next slice in the cube of brain tissue), the computer program once again colors every unique cell with a different color. However, this time, the computer program must recognize every cell in the second image that appeared in the first image and use the same colors it used in the first image.
- Finally, once all the images are colored in this manner, the last step is to stitch together the bundle of 2D images and create the 3D map by using the matching colors in each image to isolate each individual neuron and project it into 3D space.
Accuracy is incredibly important in building a 3D connectome. If you only partially color in a cell (see white arrow), then the structure of the cell in the 3D map is lost. If you fail to color the same neuron with the same color in one image in the entire cube (see black arrow), you introduce a cut in the connectivity graph.

Figure 3. The bottom left was annotated by a computer and contains mistakes. The bottom right was annotated by a professional. Source: Lichtman Lab (Harvard University)
Accuracy in image processing is more challenging with brain tissue than other types of human tissue due to these main factors: the number of types of cells (i.e. 50 in the retina alone vs. 5 in the liver), the complicated geometry of nervous tissue, the directionality of neural circuits, the fine structure of neural circuits with no structural redundancies like in the renal system, and how the cellular structure of neural tissue is a product of both genetic instruction and experience.[1:2] These are some of the reasons why even the best traditional (as opposed to deep learning) computer vision models struggle with accuracy in the task of image segmentation.
The second problem within image processing is scale. The datasets used in connectomics are some of the largest in the world. According to Lichtman et al., "acquiring images of a single cubic millimeter of a rat cortex will generate about 2 million gigabytes or 2 petabytes of data."[2:3] The volume of a rat cortex is approximately 500 mm3, which would require 1,000 petabytes of data.[2:4] To put one petabyte in context, the complete database system of Walmart is a few petabytes.[2:5] A complete human cortex is 1000 times larger than the cortex of a rat and would require processing a zettabyte of data which is about the total amount of recorded data in the entire world back in 2014.[2:6] Given that CPU and GPU computing is cost and energy-intensive, just processing this amount of data is a huge challenge, even without considering the accuracy of the output.
Up Next
To summarize, Lichtman et al. explain that the "greatest challenge facing connectomics at present is to obtain saturated reconstructions of very large (for example, 1 mm3) brain volumes in a fully automatic way, with minimal errors, and in a reasonably short amount of time".[2:7] Indeed, connectomics faces challenges on three fronts: automation, scale, and accuracy. The challenge on each of these fronts is daunting.
In part 2, we will discuss how deep learning models can help solve the accuracy and automation challenges in image processing. In contrast to traditional computer vision models, deep learning models are much more accurate, but they require more computational resources. If you enjoy building PCs, you should definitely read the next part because we discuss the precise specifications of a $1M computing system and why running deep learning models in connectomics is currently too expensive relative to traditional models.
Morgan, Joshua L., and Jeff W. Lichtman. “Why Not Connectomics?” Nature Methods 10, no. 6 (June 2013): 494–500. https://doi.org/10.1038/nmeth.2480. ↩︎ ↩︎ ↩︎
Lichtman, Jeff W, Hanspeter Pfister, and Nir Shavit. “The Big Data Challenges of Connectomics.” Nature Neuroscience 17, no. 11 (November 2014): 1448–54. https://doi.org/10.1038/nn.3837. ↩︎ ↩︎ ↩︎ ↩︎ ↩︎ ↩︎ ↩︎ ↩︎
I snapped this photo a long time ago in April 2018 when I was taking a freshman seminar called The Amazing Brain led by Prof. John Dowling. Prof. Dowling took us on a field trip to the basement of Northwest Labs to see the world's fastest (and most expensive!) scanning electron microscope. I remember being told that the microscope needed to be in the basement to minimize disruptive vibrations. The blue light glows much brighter in real life, and it makes you feel like the microscope is from the future! When I was learning about connectomics and the Lichtman lab in 2021, the mention of a new Carl Zeiss microscope in one of the articles gave me a flashback and led me to retrieve this photo. ↩︎
Manzoor, M., Singh, J., Gani, A.: Assessment of physical, microstructural, thermal, techno-functional and rheological characteristics of Apple (malus domestica) seeds of northern Himalayas. Scientific Reports. 11, (2021). https://doi.org/10.1038/s41598-021-02143-z. ↩︎
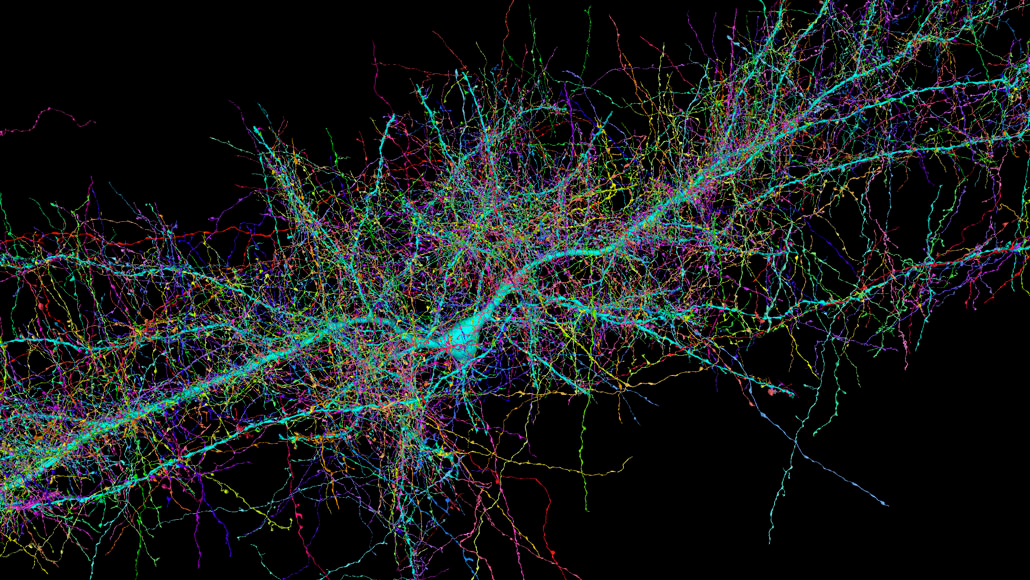
Partial Rendering of a 3D connectome
Dive Deeper

Lichtman, Jeff W, Hanspeter Pfister, and Nir Shavit. “The Big Data Challenges of Connectomics.” Nature Neuroscience 17, no. 11 (November 2014): 1448–54.
PDF: https://rdcu.be/debaO
Morgan, Joshua L., and Jeff W. Lichtman. “Why Not Connectomics?” Nature Methods 10, no. 6 (June 2013): 494–500.
PDF: https://rdcu.be/debaN
A Browsable Petascale Reconstruction of the Human Cortex. A blog post about the Google Connectomics team and the Lichtman Lab at Harvard University collaborating to create a 1.4-petabyte connectome of a 1mm3-sized piece of the human cerebral cortex (see figure 4). To browse the connectome in 3D, go to the Neuroglancer browser interface.


